6.3: Coding for Life
- Page ID
- 5663
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Scientists, Structures, and Secrets - DNA
The nucleotide bases serve as letters for our genetic code and provide the functionality of DNA and RNA. When DNA was decomposed into nucleotides, it was discovered that certain bases always appeared in the same proportions. The number of adenine and thymine nucleotides were always equal, and the number of cytosine nucleotides was the same as guanine. In 1949, Erwin Cargoff proposed base pairing would explain why nucleotides A-T and C-G always appeared in equal numbers.
Observations of DNA using x-ray crystallography further revealed the structure of the molecule. This method is akin to shining a flashlight into a hall of mirrors and determining where the mirrors are placed based on the way that the light bounces around. Rosalind Franklin had a background in physical chemistry and she had improved on x-ray crystallography techniques at the time. She produced unprecedentedly precise x-ray crystallography images while working in the lab of Maurice Wilkins. Her most famous photo held the key to DNA's structure and was known as "Photo 51." It was heralded by J.D. Bernal, the father of x-ray crystallography in biochemistry, as "among the most beautiful x-ray photographs of any substance ever taken."

Watson and Crick were theoretical biochemists. Watson discovered that the adenine-thymine bond was exactly the same length as the cytosine-guanine bond, which helped him form the picture of each base pair as rungs of a helical ladder. Francis Crick helped to develop a mathematical model for the diffraction pattern of a helical structure. In 1951, Crick and Watson began to work together and when Maurice Wilkins showed them Rosalind Franklin's Photo 51, they were able to piece together the double-helix model of DNA.
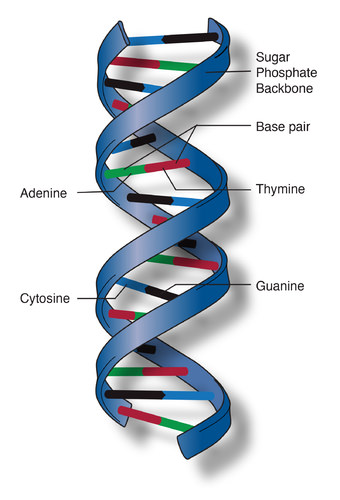
The double-helix of DNA is composed of the sugar and phosphate components of nucleotides. The bases stick out from this backbone and bind to their appropriate counterpart through relatively weak hydrogen bonds. In the most common form, the bases appear parallel to each other, like a well-designed stairwell. A slight electrostatic repulsion helps to support this structure. This structure is stable in aqueous environments because the hydrophobic base pairs are protected by the sugar and phosphate backbone. The double-stranded nature of DNA affords a rigid, stable, and long-lived structure. During DNA replication, each strand is checked against the other to reduce copying errors or accidental mutations.
The Genetic Code
DNA encodes and stores genes, which provide a very lengthy instructions manual for all living creatures on Earth. If you took the DNA in one human cell and completely stretched it out, it would measure roughly 2 meters, the average height of an NBA player. If you took all of the DNA from all of the cells in a human body and joined them end-to-end, they would cross the diameter of the Solar System twice.
Different segments of DNA are known as chromosomes. An organism's genome is the complete collection of chromosomes. Humans have 23 pairs of chromosomes, encoding roughly 25,000 genes using about 3 billion base pairs. A mosaic of the entire human genome was sequenced between 1990 and 2008 in twenty different institutions in six different countries. This remains one of the most impressive collaborative projects in science.
| Species | Base Pairs | Genes |
| Virus | 170,000 | ? |
| E. Coli | 4,600,000 | 3,200 |
| Fruit Fly | 180,000,000 | 13,600 |
| Chicken | 1,000,000,000 | 23,000 |
| Corn | 2,500,000,000 | 59,000 |
| Human | 3,000,000,000 | 25,000 |
| Lily | 100,000,000,000 | ? |
| Grasshopper | 180,000,000,000 | ? |
| Amoeba | 670,000,000,000 | ? |
My, What Big Genomes You Have
All the better to encode with... or is it? It seems intuitive that a larger genome should corresponds to more complex organisms, but that would be wrong. An example of different genome sizes is given by the table above; humans have longer genomes than chickens, but we lose out to corn.
Most DNA is known as noncoding DNA and does not translate directly to genes. It is used instead to signal the start of a gene, to help with DNA coiling, and potentially to carry out several other functions that we have yet to discover. Remarkably, more than 98% of the human genome is non-coding. In contrast, only 20% of the DNA in bacteria is noncoding DNA. The bladderwort plant currently holds the record for most efficient genome with only 3% noncoding DNA.
DNA encodes important instructions for life, but it can become damaged when base pairs or whole segments of DNA are deleted, inverted, duplicated, or moved around. Mutations can be damaging, for example, causing cells to become cancerous. Damage sustained to the phosphate-sugar backbone of DNA is one of the primary causes of mutations and can be a result of exposure to UV radiation. For this reason, sunscreen remains a multi-million dollar industry.
However, mutations can also occur naturally, resulting in expressed altered genes that give rise to new characteristics. This is the mechanism for Darwinian evolution: beneficial traits arising from mutation will be preferentially selected when mating and propagated through succeeding generations. Natural mutations arise at measurable rates for different species. This mutation rate allows us to measure the genetic distance between species. This value is obtained by determining the statistical number of mutations required to change one species' genome into another's. For example, deer and giraffes are close in genetic difference. But, both deer and giraffes are genetically quite far from sunflowers. A genome of a species helps to reveal its evolutionary path.
Question for Thought
If the history of an organism can be traced through their genetic code, is it possible to trace this history all the way back to the beginning of life
Translating the Genetic Code
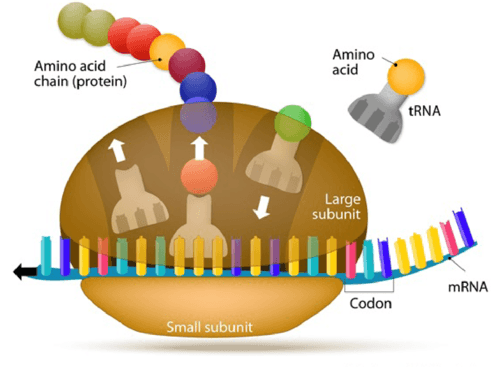
The genetic code in DNA is used to manufacture proteins with the help of RNA. Messenger RNA (mRNA) transcribes the code from the DNA and carries this information to the ribosome (the protein factories of the cell). A ribosome will read the nucleotide code from mRNA and translate it to build amino acids. Each amino acid is specified by different codons, a sequence of three base pairs. Transfer RNA (tRNA) matches up each codon with the appropriate amino acid. The resulting polypeptide chain can then be folded into the needed proteins.
RNA World
How did this amazing biochemical system begin and how did it evolve? Proteins are needed to catalyze chemical reactions critical to the survival of cells. But, it is difficult to imagine how proteins could have been the precursor to living cells because DNA is required to manufacture proteins, presenting a "chicken-and-egg" problem. A breakthrough came when Sidney Altman and Thomas Czech discovered a class of RNA molecules called ribozymes that could catalyze their own replication. Ribozymes show that RNA, which can encode genetic information, can also act as an enzyme. This discovery was awarded the Nobel prize in 1989 and supported a hypothesis called the Early RNA world, where ancient life used RNA for storing genetic information and catalyzing chemical reactions. This hypothesis had been suggested in the 1960's by Carl Woese, Frances Crick and Leslie Orgel. According to this hypothesis, the instability of RNA promoted mutations and natural selection eventually evolved a more stable, double-stranded DNA molecule as ribozymes were phased out. A fascinating "smoking gun" for this hypothesis is the fact that the ribosome, which assembles proteins in cells today, is a ribozyme. While the current day ribosome incorporates some proteins, none of the proteins are anywhere near the active site where the reactions take place. They appear to exist largely for structural support.

While the capabilities of RNA seem to make it the perfect candidate to explain the origin of life, RNA is a far more complicated structure and not as easy to make as amino acids. Miller-Urey experiments have been capable of synthesizing a series of smaller, very reactive molecules. When enough of these molecules are made, detectable amounts of purine and pyrimidine bases can be detected. Components of nucleotides have also been discovered on meteorites like the Murchison Meteorite. However, no complete extraterrestrial nucleotides or nucleic acid chains have been discovered yet.
A breakthrough came in 2009 from British chemist John Sutherland along with Matthew Powner and Beatrice Gerland. Sutherland's group determined a chemically efficient pathway for nucleotides to form that are plausible in a prebiotic environment. Rather than form each component of the nucleotide individually, they proposed a method that formed and attached a purine and a ribose sugar in the same reaction. Phosphate is used to help catalyze the reaction and incorporated into the nucleotide later.
Once nucleotides are assembled, perhaps they themselves could assemble to form RNA. RNA is capable of assembling proteins, and proteins run the reactions that organize life. Is that all there is to life then? At what point does a collection of molecules and equilibrium reactions become a living organism?
Biochemistry to Biology
What is life? This is a hard enough question for philosophers to answer, but perhaps even harder to answer scientifically. Biologists can distinguish smaller and smaller structures and biochemists are reaching a greater understanding of the chemistry of life. But, there is still a wide gap in our understanding. How do we bridge our understanding of chemistry with our understanding of living cells?
An over-arching, all-encompassing, definition of life currently eludes us. The working definition used within NASA is that life is "a self-sustaining chemical system capable of Darwinian evolution." Life is a chemical system: CHON, amino acids, nucleotides. Life is self-sustaining: life generates its own energy for reactions and so must also maintain reactions that generate energy. Life is capable of Darwinian evolution: an imperfect genetic system stores the information that defines an organism, and also suffers from errors that give rise to mutations and new characteristics. Living organisms reproduce and are capable of adapting and changing to become better suited for different environments.
Membranes
Cells are bound and regulated by membranes. Membranes are composed of of phospholipids with of a hydrophilic "head" that are water soluble and two hydrophobic "tails", which try to avoid interacting with water. It is not clear how the first phospholipids formed, but it is possible that the earliest membranes were first composed of a less complicated lipid connected to hydrocarbon chains.
Phospholipids assembled into bilayer sheets allow cell membranes to be both fluid and semi-permeable. Cell membranes are fluid in the sense that the different phospholipids can move around past each other, which allow proteins that are incorporated into the membrane to be inserted and similarly move around. Cell membranes are also semi-permeable, allowing only certain substances through. Most molecules cannot spontaneously diffuse though both a polar and nonpolar region. Substances can therefore only cross the phospholipid bilayer if there is significant pressure (either physical or chemical) driving diffusion or they have some help. To this end, cell membranes incorporate several proteins specially designed to control the traffic of molecules in and out of the cell. Membranes provide a self-contained environment to safeguard the reactions that drive life.

The Last Universal Common Ancestor
The Last Universal Common Ancestor (LUCA) on Earth is a concept, rather than an actual organism. Any universal characteristics of life on Earth are either universal because they are inherited or because they are truly fundamental to life in general. Without a second example of life we are unable to understand how common these features will be on other worlds.
LUCA represents the earliest shared qualities of ancestral life and likely appeared on Earth between 3.5 to 4 Gya and seeded our planet with life. All of the features of life today would have been inherited from LUCA. From what we know about terrestrial biology, this means that LUCA would have been carbon-based, dependent on water, incorporated left-handed amino acids, and used ATP for energy transport. LUCA would have used DNA or RNA to encode genes and translate them into proteins. Significantly, the codons that translate for specific amino acids are the same in every known organism. This code must have been passed down from a common ancestor, from which every other species has since evolved. LUCA might not have even been as sophisticated as a single celled organism.
One can only imagine how the field of biology will change the moment extraterrestrial life is discovered for the first time. Will we see shocking similarities? Or will life elsewhere be so different that it is barely recognizable to us?

